The AnyMoCap Framework#
The AnyMoCap model is an effort to create a simple and unified framework for doing any kind of MoCap analysis with the AnyBody Modeling System.
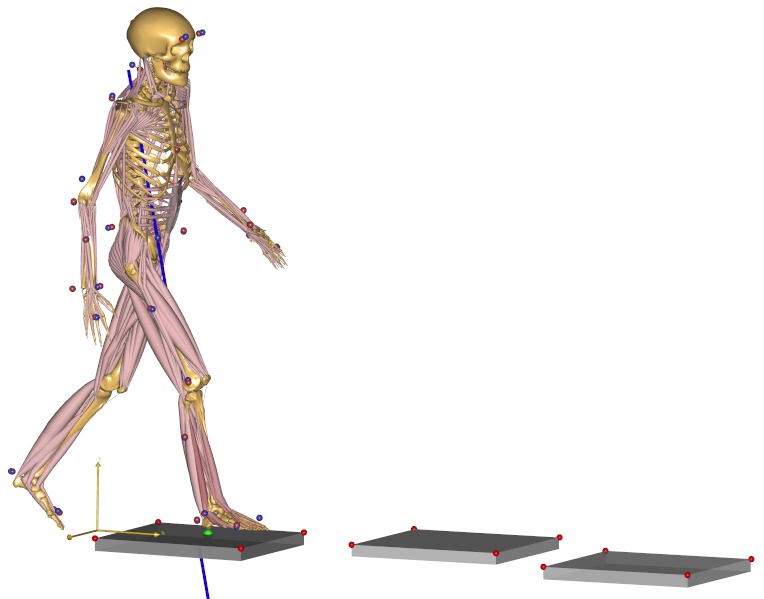
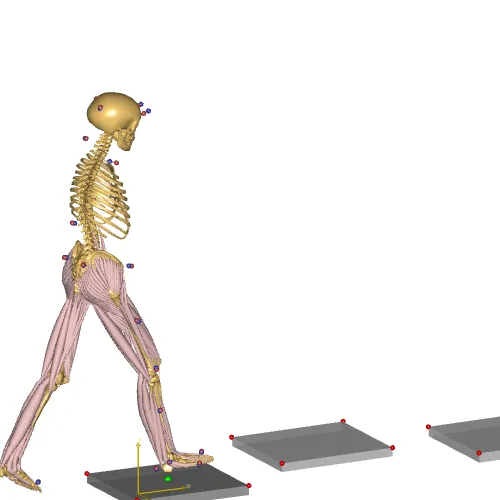
Fig. 9 Making MoCap models the easier#
Features#
Adapts to any Mocap protocol
Optimization of marker locations and anthropometrics.
Support Marker based (C3D) and inertial based (BVH) input.
Support for standard force plates: (Types 1-4) plus more.
Easy setup with multiple trials and subjects
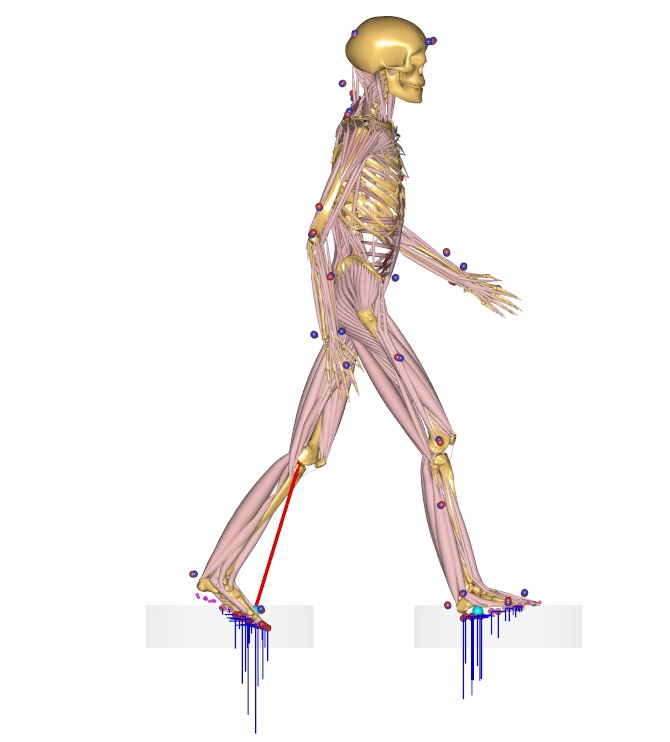
Over determinate kinematic analysis#
See also
The AnyBody tutorials, and the lesson on using the AnyMoCap model
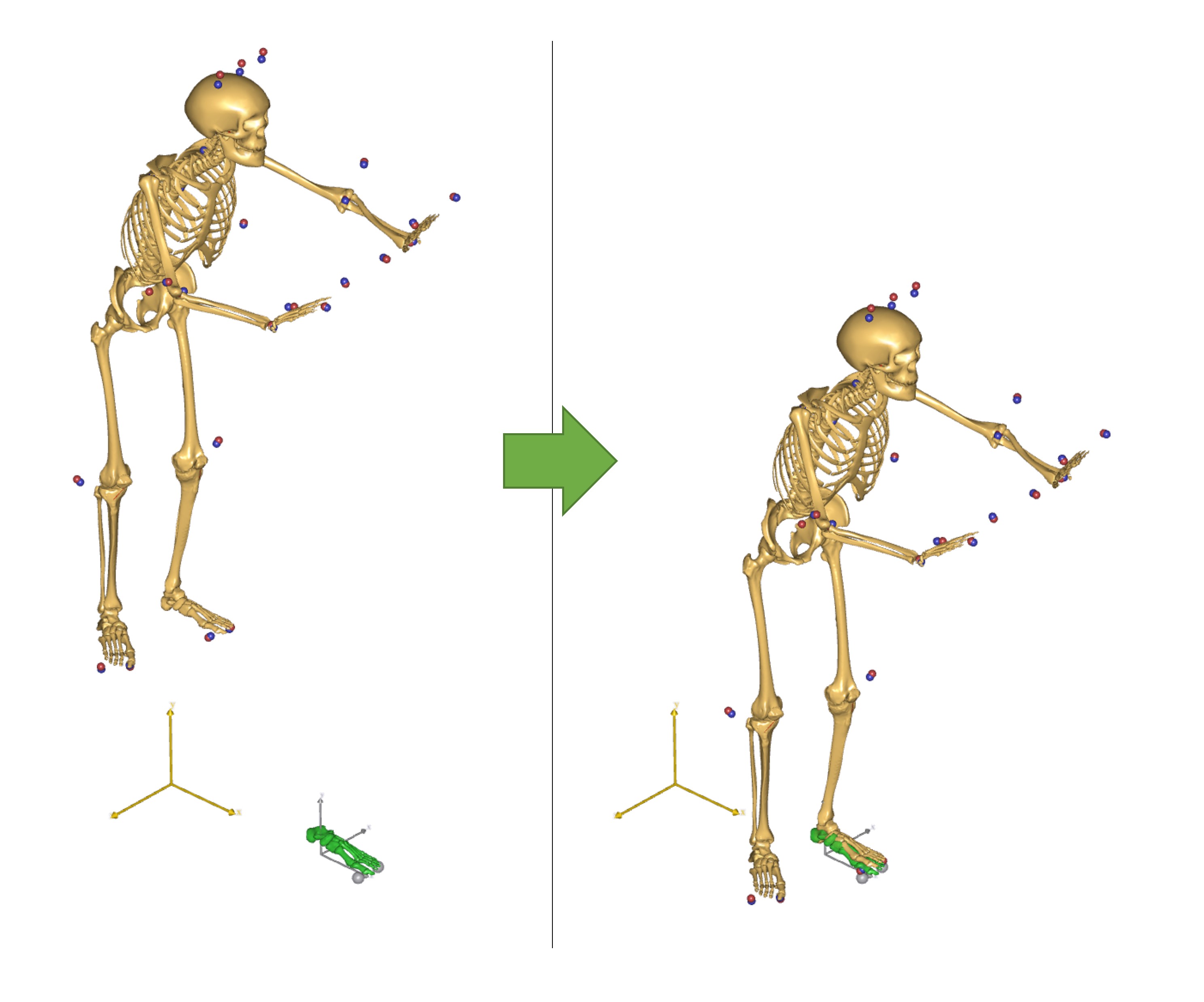
Musculoskeletal models that use Motion capture data are different from other types of models found the AMMR. Most importantly, MoCap models usually require an over-determinate kinematic solver to handle the excess in information that the optical markers provide.
The over-determinate solver in AMS works great, but it can only find velocities and accelerations numerically. That has some performance issue when running inverse dynamics analysis. To overcome the problem, the MoCap analysis is split into a two-step procedure, as illustrated on the figure below:
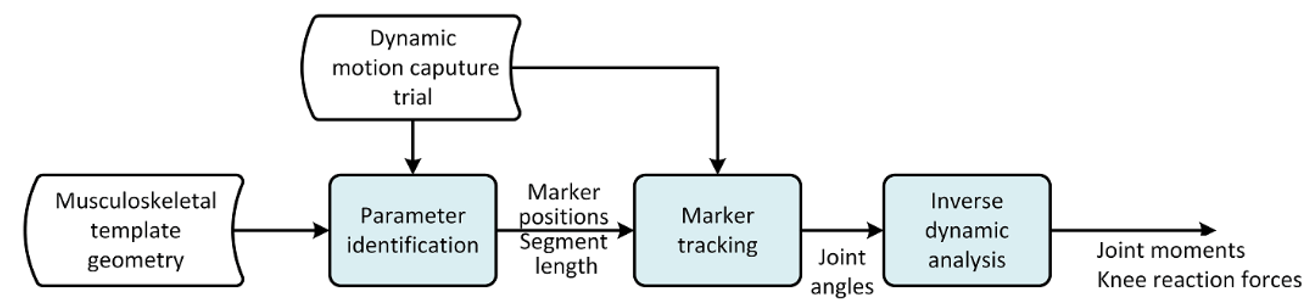
Fig. 10 Figure 1: The framework of the AnyMoCap model. Analysis is split into three steps. Parameter identification, Marker tracking, Inverse dynamic analysis.#
The over determinate kinematic analysis solves the model for positions, and writes joint angles to a a set of files. These joint angles can then be used with the determinate kinematic solver in the inverse dynamic analysis.
AnyMoCap examples#
The following gallery shows different examples using the AnyMoCap framework.
Tip
The Multi trial MoCap model is likely the best starting point when working with many trials/subjects from a MoCap experiment.
Get started#
Caution
It is recommended not to place working models inside the AMMR folder. Copy the example elsewhere before using.
The easiest way to get started, is to adapt one the example application above.
A typical application using the AnyMoCap framework will look something like this:
1 #include "../libdef.any"
2
3 #path MOCAP_TRIAL_SPECIFIC_DATA "TrialSpecificData.any"
4 #path MOCAP_SUBJECT_SPECIFIC_DATA "SubjectSpecificData.any"
5 #path MOCAP_LAB_SPECIFIC_DATA "LabSpecificData.any"
6
7 // Include the AnyMoCap Framwork
8 #include "<ANYMOCAP_MODEL>"
You can place this main file anywhere on your computer as long the
libdef.any file points to model repository (AMMR) you want to use.
Model structure#
When loading the AnyMoCap model the following layout shows up in the model tree:
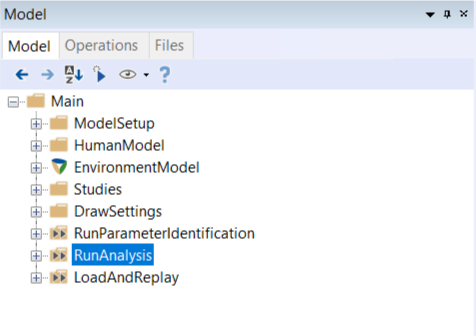
Description of Model folders#
Options and settings#
The Main.ModelSetup folder contains the main machinery of the AnyMoCap
framework. It is controlled by a number of settings both #define/#path switches
and variables which can be set directly. For example:
#define MOCAP_INPUT_DATA_TYPE "C3D"
Main.ModelSetup.TrialSpecificData.LastFrame = 240;
The following two sections gives an overview of the settings available:
Paths settings and switches#
Path settings and switches are usually prefixed by MOCAP_ to indicate that
they are specific to the AnyMoCap framework.
Configurable variables#
The Main.ModelSetup folder contains all the machinery of the AnyMoCap
framework. It has three important subfolder (TrialSpecificData,
SubjectSpecificData and LabSpecificData).
Fig. 11 These three folders contain variables specific to the application but also has a few variables which can be assigned directly by the user.#
More resources#
This section contains further documentation on the AnyMoCap framework.